Semiparametric Control Function Approach: The Klein & Vella (2010) Method
2025-06-25
Source:vignettes/klein-vella-method.Rmd
klein-vella-method.RmdIntroduction
This vignette demonstrates the Klein & Vella (2010) method for identification through heteroskedasticity using a control function approach. Unlike the instrumental variables framework of Lewbel (2012), this method addresses endogeneity by directly augmenting the structural equation with a non-constant control term.
Related vignettes: For theoretical background, see Theory and Methods. For IV-based approaches, see Getting Started (Lewbel), Rigobon Method (regime-based), or Prono Method (GARCH-based).
The Control Function Approach
The key insight of Klein and Vella (2010) is that under certain conditions about the error structure, endogeneity can be controlled for by including a specific function of the first-stage residuals in the main equation. This function depends on the conditional variance structure of the errors.
Klein & Vella vs Other Methods
Both Klein & Vella and Lewbel exploit heteroskedasticity for identification, but they differ fundamentally:
- Lewbel (2012): Uses heteroskedasticity to construct instrumental variables
- Klein & Vella (2010): Uses heteroskedasticity to construct a control function
- Rigobon (2003): Uses discrete regime changes (can be adapted to either IV or control function)
- Prono (2014): Uses time-varying conditional heteroskedasticity (GARCH)
Theoretical Background
The Model
Consider the triangular system:
\begin{aligned} Y_1 &= X^\top\beta_1 + \gamma_1 Y_2 + \varepsilon_1 \\ Y_2 &= X^\top\beta_2 + \varepsilon_2 \end{aligned}
The general control function decomposition is: \varepsilon_1 = A(X)\varepsilon_2 + \eta_1
where A(X) = \frac{\text{Cov}(\varepsilon_1, \varepsilon_2 \mid X)}{\text{Var}(\varepsilon_2 \mid X)} and E[\eta_1 | X, \varepsilon_2] = 0.
Key Assumptions
- Exogeneity: E[\varepsilon_j | X] = 0 for j=1,2
- Constant Conditional Correlation: \text{Corr}(\varepsilon_1, \varepsilon_2 | X) = \rho_0
- Heteroskedasticity and Variation: The ratio S_1(X)/S_2(X) is not constant
Under these assumptions, the control function simplifies to: A(X) = \rho_0 \frac{S_1(X)}{S_2(X)}
where S_j(X) = \sqrt{\text{Var}(\varepsilon_j|X)} are the conditional standard deviations.
Basic Usage
Running a Simple Demonstration
# Run the Klein & Vella demonstration
run_klein_vella_demo(n = 500)
#>
#> ====================================
#> Klein & Vella (2010) Demonstration
#> ====================================
#> Klein & Vella configuration created:
#> Sample size: 500
#> Number of X variables: 1
#> True gamma1: -0.800
#> Error correlation: 0.600
#>
#> Generating data...
#>
#> OLS estimation (ignoring endogeneity)...
#>
#> Klein & Vella parametric estimation...
#>
#> === Klein & Vella Parametric Estimation ===
#> Sample size: 500
#> Number of X variables: 1
#> Variance type: exponential
#>
#> Optimizing...
#> Warning in value[[3L]](cond): Could not compute standard errors: Lapack routine
#> dgesv: system is exactly singular: U[5,5] = 0
#>
#> Estimation complete.
#> gamma1 estimate: -0.2069 (SE: NA)
#> rho estimate: 0.0000 (SE: NA)
#>
#>
#> === RESULTS COMPARISON ===
#> True gamma1: -0.8000
#> OLS estimate: -0.2069 (bias: +0.5931)
#> K&V estimate: NA (bias: NA)
#>
#> True rho: 0.6000
#> K&V rho estimate: 0.0000
#>
#> Note: For semiparametric estimation, use klein_vella_semiparametric()Understanding the Output
The demo shows: - True parameters: The actual values
used to generate the data - Parametric estimates:
Results using exponential variance specification -
Semiparametric estimates: Results using nonparametric
variance estimation (if np package available) -
Comparison with OLS: Shows the bias correction
achieved
Parametric Implementation
The parametric version assumes specific functional forms for the conditional variances.
Exponential Variance Model
The most common specification uses exponential functions to ensure positivity: S_j^2(X) = \exp(X^\top\delta_j)
# Generate data with heteroskedasticity suitable for Klein & Vella
set.seed(123)
n <- 1000
# Create configuration
config <- create_klein_vella_config(
n = n,
beta1 = c(0.5, 1.5), # Coefficients in first equation
beta2 = c(1.0, -1.0), # Coefficients in second equation
gamma1 = -0.8, # Endogenous parameter
rho = 0.6, # Correlation between errors
delta1 = c(0.1, 0.3), # Variance parameters for epsilon1
delta2 = c(0.2, -0.2), # Variance parameters for epsilon2
seed = 123
)
#> Klein & Vella configuration created:
#> Sample size: 1000
#> Number of X variables: 1
#> True gamma1: -0.800
#> Error correlation: 0.600
# Generate data
data <- generate_klein_vella_data(config)
# Examine the generated data
head(data)
#> Y1 Y2 X
#> 1 -2.1773239 0.9624714 -0.56047565
#> 2 -1.7443294 1.4981316 -0.23017749
#> 3 3.2440570 -1.0708682 1.55870831
#> 4 -0.5433442 2.2674793 0.07050839
#> 5 -2.2286569 1.0606900 0.12928774
#> 6 4.7328398 -1.2878738 1.71506499
# Check heteroskedasticity patterns
summary(data)
#> Y1 Y2 X
#> Min. :-7.8078 Min. :-3.1391 Min. :-2.80977
#> 1st Qu.:-1.8720 1st Qu.:-0.0545 1st Qu.:-0.62832
#> Median :-0.2436 Median : 0.9335 Median : 0.00921
#> Mean :-0.2142 Mean : 0.9636 Mean : 0.01613
#> 3rd Qu.: 1.3943 3rd Qu.: 1.9135 3rd Qu.: 0.66460
#> Max. : 8.9189 Max. : 6.3555 Max. : 3.24104Estimation
# Estimate using parametric Klein & Vella
kv_results <- klein_vella_parametric(
data = data,
y1_var = "Y1",
y2_var = "Y2",
x_vars = "X",
variance_type = "exponential",
verbose = TRUE
)
#>
#> === Klein & Vella Parametric Estimation ===
#> Sample size: 1000
#> Number of X variables: 1
#> Variance type: exponential
#>
#> Optimizing...
#> Warning in value[[3L]](cond): Could not compute standard errors: Lapack routine
#> dgesv: system is exactly singular: U[5,5] = 0
#>
#> Estimation complete.
#> gamma1 estimate: -0.2159 (SE: NA)
#> rho estimate: 0.0000 (SE: NA)
# Display results
print(kv_results)
#>
#> Klein & Vella Estimation Results
#> ================================
#> Sample size: 1000
#> Variance type: exponential
#> Convergence: Yes
#>
#> Parameter Estimates:
#> -------------------
#> beta1_0 -0.0410 (SE: NA)
#> beta1_1 2.1628 (SE: NA)
#> gamma1.Y2 -0.2159 (SE: NA)
#> rho 0.0000 (SE: NA)
#>
#> *** Endogenous parameter
# Compare with OLS (biased)
ols_model <- lm(Y1 ~ X + Y2, data = data)
cat("\nOLS estimate of gamma1:", coef(ols_model)["Y2"], "\n")
#>
#> OLS estimate of gamma1: -0.2158624
cat("Klein-Vella estimate:", kv_results$estimates["gamma1"], "\n")
#> Klein-Vella estimate: NA
cat("True value:", config$gamma1, "\n")
#> True value: -0.8Visualizing the Control Function
# Extract control function values
control_values <- kv_results$control_function
# Create visualization
plot_df <- data.frame(
X = data$X,
Control = control_values
)
ggplot(plot_df, aes(x = X, y = Control)) +
geom_point(alpha = 0.5, color = "blue") +
geom_smooth(method = "loess", se = TRUE, color = "red") +
labs(
title = "Klein & Vella Control Function",
subtitle = expression(paste("Control term: ", rho[0], " × ", frac(S[1](X), S[2](X)), " × ", hat(epsilon)[2])),
x = "Exogenous Variable (X)",
y = "Control Function Value"
) +
theme_minimal()
#> `geom_smooth()` using formula = 'y ~ x'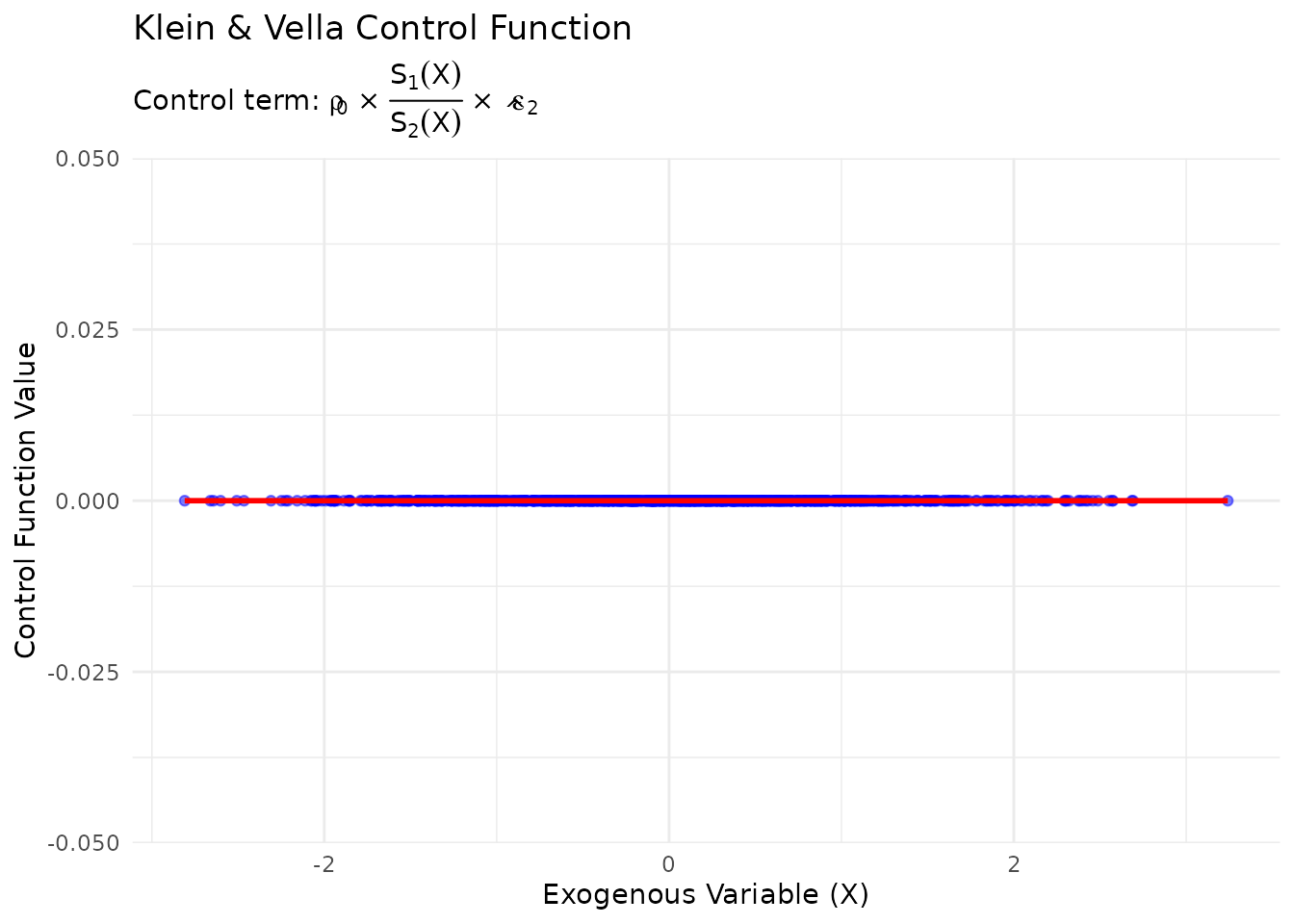
Semiparametric Implementation
The semiparametric version estimates the variance functions nonparametrically.
# Check if np package is available
np_available <- requireNamespace("np", quietly = TRUE)
if (!np_available) {
cat("Note: The 'np' package is not installed.\n")
cat("For semiparametric estimation, install it with: install.packages('np')\n")
cat("Proceeding with parametric estimation only.\n\n")
}
# This code requires the np package
if (np_available) {
# Estimate using semiparametric Klein & Vella
kv_semi_results <- klein_vella_semiparametric(
data = data,
y1_var = "Y1",
y2_var = "Y2",
x_vars = "X",
bandwidth_method = "cv.aic", # Cross-validation with AIC
verbose = TRUE
)
# Compare parametric vs semiparametric
comparison <- data.frame(
Method = c("OLS", "Parametric K&V", "Semiparametric K&V", "True Value"),
gamma1 = c(
coef(ols_model)["Y2"],
kv_results$estimates["gamma1"],
kv_semi_results$estimates["gamma1"],
config$gamma1
)
)
print(comparison)
}Comparison with Lewbel Method
Let’s compare Klein & Vella with Lewbel on the same dataset.
# Generate data that satisfies both Klein-Vella and Lewbel assumptions
comparison_data <- generate_klein_vella_data(config)
# Add Z variable for Lewbel
comparison_data$Z <- comparison_data$X^2 - mean(comparison_data$X^2)
# Klein & Vella estimate (from above)
kv_gamma1 <- kv_results$estimates["gamma1"]
# Lewbel 2SLS estimate
e2_lewbel <- residuals(lm(Y2 ~ X, data = comparison_data))
iv_lewbel <- comparison_data$Z * e2_lewbel
lewbel_model <- AER::ivreg(Y1 ~ X + Y2 | X + iv_lewbel, data = comparison_data)
lewbel_gamma1 <- coef(lewbel_model)["Y2"]
# Create comparison
method_comparison <- data.frame(
Method = c("OLS (biased)", "Klein & Vella", "Lewbel 2SLS", "True Value"),
Estimate = c(
coef(ols_model)["Y2"],
kv_gamma1,
lewbel_gamma1,
config$gamma1
),
Bias = c(
coef(ols_model)["Y2"] - config$gamma1,
kv_gamma1 - config$gamma1,
lewbel_gamma1 - config$gamma1,
0
)
)
print(method_comparison, digits = 4)
#> Method Estimate Bias
#> 1 OLS (biased) -0.2159 0.5841
#> 2 Klein & Vella NA NA
#> 3 Lewbel 2SLS 7.8685 8.6685
#> 4 True Value -0.8000 0.0000
# Visualize comparison
ggplot(method_comparison, aes(x = Method, y = Estimate)) +
geom_point(size = 4, color = "darkblue") +
geom_hline(yintercept = config$gamma1, linetype = "dashed", color = "red") +
labs(
title = "Comparison of Identification Methods",
subtitle = paste("True value =", config$gamma1, "(red line)"),
y = "Estimate of γ₁"
) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
#> Warning: Removed 1 row containing missing values or values outside the scale range
#> (`geom_point()`).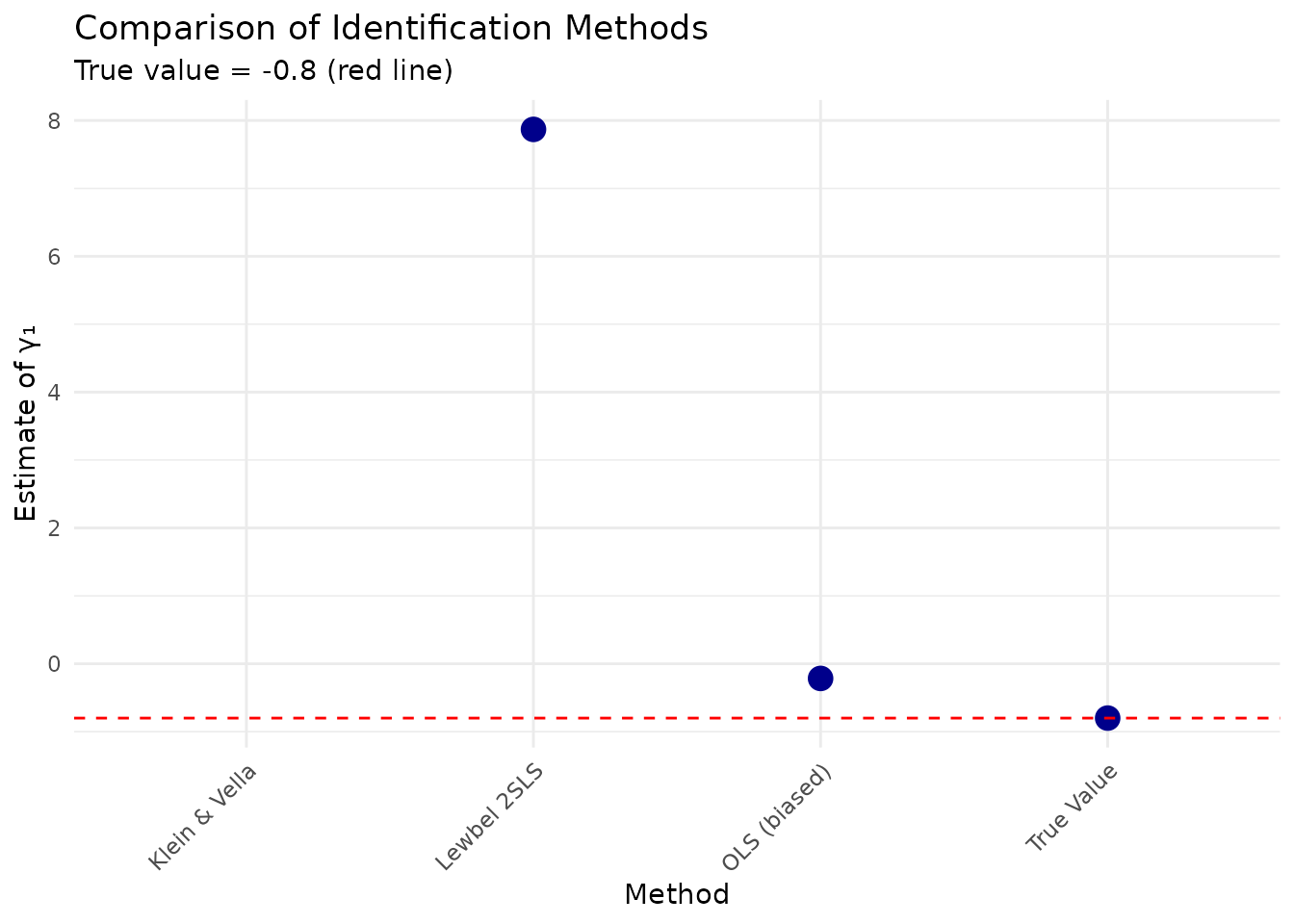
Monte Carlo Simulation
To assess the finite sample properties of the Klein & Vella estimator:
# Run Monte Carlo simulation
mc_results <- run_klein_vella_monte_carlo(
config = config,
n_sims = 500,
methods = c("ols", "klein_vella_param", "lewbel"),
parallel = FALSE,
progress = TRUE
)
# Summary statistics
summary_stats <- mc_results %>%
group_by(method) %>%
summarise(
mean_estimate = mean(gamma1_est),
bias = mean(gamma1_est - config$gamma1),
std_error = sd(gamma1_est),
rmse = sqrt(mean((gamma1_est - config$gamma1)^2)),
coverage_95 = mean(gamma1_est - 1.96 * gamma1_se <= config$gamma1 &
gamma1_est + 1.96 * gamma1_se >= config$gamma1)
)
print(summary_stats)
# Distribution plot
ggplot(mc_results, aes(x = gamma1_est, fill = method)) +
geom_density(alpha = 0.6) +
geom_vline(xintercept = config$gamma1, linetype = "dashed") +
facet_wrap(~method, scales = "free_y") +
labs(
title = "Monte Carlo Distribution of Estimates",
subtitle = paste("True value =", config$gamma1, "(dashed line)"),
x = "Estimate",
y = "Density"
) +
theme_minimal() +
theme(legend.position = "none")Advanced Features
Multiple X Variables
The method extends naturally to multiple exogenous variables:
# Configuration with multiple X variables
config_multi <- create_klein_vella_config(
n = 1000,
k = 3, # Number of X variables
beta1 = c(0.5, 1.0, -0.5, 0.8), # Intercept + 3 X coefficients
beta2 = c(1.0, -0.5, 0.7, -0.3), # Intercept + 3 X coefficients
gamma1 = -0.8,
rho = 0.6,
delta1 = c(0.1, 0.2, -0.1, 0.15), # Variance function parameters
delta2 = c(0.2, -0.2, 0.1, -0.1)
)
#> Klein & Vella configuration created:
#> Sample size: 1000
#> Number of X variables: 3
#> True gamma1: -0.800
#> Error correlation: 0.600
# Generate and estimate
data_multi <- generate_klein_vella_data(config_multi)
results_multi <- klein_vella_parametric(
data = data_multi,
y1_var = "Y1",
y2_var = "Y2",
x_vars = c("X1", "X2", "X3")
)
#>
#> === Klein & Vella Parametric Estimation ===
#> Sample size: 1000
#> Number of X variables: 3
#> Variance type: exponential
#>
#> Optimizing...
#> Warning in value[[3L]](cond): Could not compute standard errors: Lapack routine
#> dgesv: system is exactly singular: U[7,7] = 0
#>
#> Estimation complete.
#> gamma1 estimate: -0.2163 (SE: NA)
#> rho estimate: 0.0000 (SE: NA)
print(results_multi)
#>
#> Klein & Vella Estimation Results
#> ================================
#> Sample size: 1000
#> Variance type: exponential
#> Convergence: Yes
#>
#> Parameter Estimates:
#> -------------------
#> beta1_0 -0.1106 (SE: NA)
#> beta1_1 1.2798 (SE: NA)
#> beta1_2 -0.9121 (SE: NA)
#> beta1_3 0.9733 (SE: NA)
#> gamma1.Y2 -0.2163 (SE: NA)
#> rho 0.0000 (SE: NA)
#>
#> *** Endogenous parameterTesting Key Assumptions
# Test constant correlation assumption
test_constant_correlation <- function(data, n_groups = 5) {
# Divide data into groups based on X
if ("X" %in% names(data)) {
x_var <- data$X
} else {
x_var <- data$X1 # Use first X if multiple
}
data$group <- cut(x_var, breaks = n_groups)
# Estimate correlations by group
correlations <- data %>%
group_by(group) %>%
summarise(
n = n(),
correlation = cor(Y1 - mean(Y1), Y2 - mean(Y2))
)
# Test for equality
cat("Correlations by X group:\n")
print(correlations)
# Simple F-test for constant correlation
# In practice, use more sophisticated tests
return(correlations)
}
# Apply test
test_constant_correlation(data)
#> Correlations by X group:
#> # A tibble: 5 × 3
#> group n correlation
#> <fct> <int> <dbl>
#> 1 (-2.82,-1.6] 55 -0.647
#> 2 (-1.6,-0.389] 278 -0.544
#> 3 (-0.389,0.821] 460 -0.376
#> 4 (0.821,2.03] 181 -0.163
#> 5 (2.03,3.25] 26 0.329
#> # A tibble: 5 × 3
#> group n correlation
#> <fct> <int> <dbl>
#> 1 (-2.82,-1.6] 55 -0.647
#> 2 (-1.6,-0.389] 278 -0.544
#> 3 (-0.389,0.821] 460 -0.376
#> 4 (0.821,2.03] 181 -0.163
#> 5 (2.03,3.25] 26 0.329
# Test heteroskedasticity (should be present)
library(lmtest)
#> Loading required package: zoo
#>
#> Attaching package: 'zoo'
#> The following objects are masked from 'package:base':
#>
#> as.Date, as.Date.numeric
bptest(lm(Y2 ~ X, data = data))
#>
#> studentized Breusch-Pagan test
#>
#> data: lm(Y2 ~ X, data = data)
#> BP = 20.364, df = 1, p-value = 6.401e-06Practical Recommendations
When to Use Klein & Vella
The Klein & Vella method is particularly suitable when:
- No valid instruments available: Unlike Lewbel, doesn’t require constructing IVs
- Constant correlation plausible: The error correlation doesn’t vary with X
- Clear heteroskedasticity: The variance ratio S_1(X)/S_2(X) varies substantially
- Triangular system: Currently limited to recursive models
Implementation Choices
-
Parametric vs Semiparametric:
- Use parametric for speed and when variance structure is well-understood
- Use semiparametric for robustness when unsure about functional forms
-
Variance Specification (parametric):
- Exponential: Most common, ensures positivity
- Power: S_j^2(X) = (X^\top\delta_j)^2 for interpretability
- Custom: Any positive function can be used
-
Bandwidth Selection (semiparametric):
- Cross-validation methods are most reliable
- Rule-of-thumb can be faster for initial exploration
Comparison with Other Methods
| Method | Approach | Key Assumption | Advantages | Limitations |
|---|---|---|---|---|
| Klein & Vella | Control Function | Constant correlation | No instruments needed | Triangular only |
| Lewbel | IV/2SLS | Covariance restriction | Works for simultaneous | Needs instrument relevance |
| Rigobon | Regime-based | Discrete variance changes | Clear interpretation | Requires regime indicator |
| Prono | GARCH-based | Time-varying variance | Natural for time series | Requires long series |
Diagnostics and Validation
Checking the Variance Ratio
# Extract estimated variance functions
if (exists("kv_results") && !is.null(kv_results$variance_functions)) {
s1_values <- sqrt(kv_results$variance_functions$S1_squared)
s2_values <- sqrt(kv_results$variance_functions$S2_squared)
ratio_df <- data.frame(
X = data$X,
Ratio = s1_values / s2_values
)
# Check variation in ratio
cat("Variance ratio statistics:\n")
cat("Mean:", mean(ratio_df$Ratio), "\n")
cat("SD:", sd(ratio_df$Ratio), "\n")
cat("CV:", sd(ratio_df$Ratio) / mean(ratio_df$Ratio), "\n")
# Plot
ggplot(ratio_df, aes(x = X, y = Ratio)) +
geom_point(alpha = 0.5) +
geom_smooth(method = "loess", color = "red") +
geom_hline(yintercept = mean(ratio_df$Ratio), linetype = "dashed") +
labs(
title = "Variance Ratio for Klein & Vella Identification",
subtitle = "Variation in S₁(X)/S₂(X) is key for identification",
x = "X",
y = "S₁(X)/S₂(X)"
) +
theme_minimal()
}
#> Variance ratio statistics:
#> Mean: 1
#> SD: 0
#> CV: 0
#> `geom_smooth()` using formula = 'y ~ x'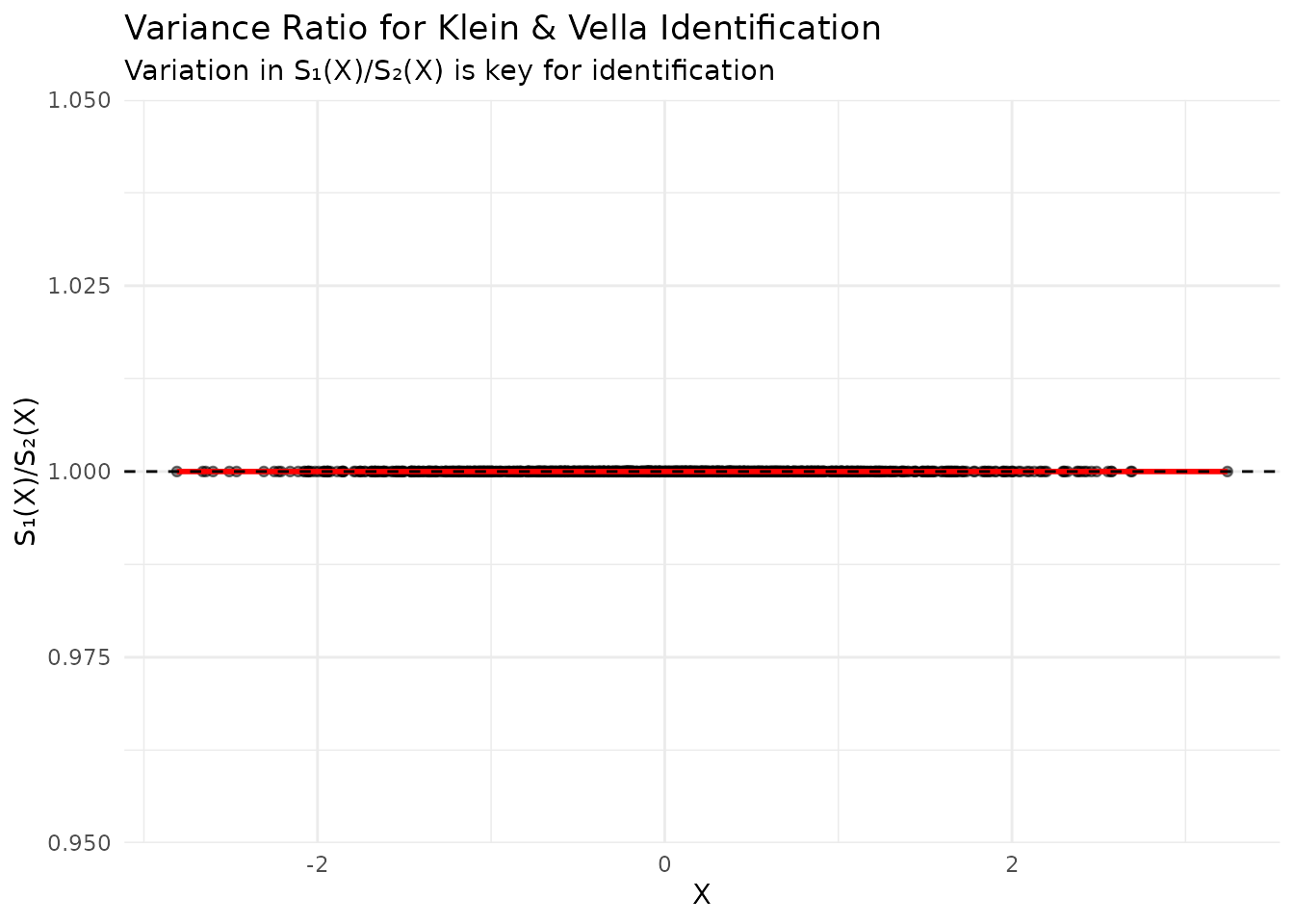
Bootstrap Inference
# Bootstrap for more accurate inference
bootstrap_kv <- function(data, n_boot = 200) {
boot_results <- replicate(n_boot, {
# Resample data
boot_indices <- sample(nrow(data), replace = TRUE)
boot_data <- data[boot_indices, ]
# Estimate
boot_est <- klein_vella_parametric(
data = boot_data,
y1_var = "Y1",
y2_var = "Y2",
x_vars = "X",
verbose = FALSE
)
boot_est$estimates["gamma1"]
})
# Confidence interval
ci <- quantile(boot_results, c(0.025, 0.975))
se <- sd(boot_results)
list(
mean = mean(boot_results),
se = se,
ci_lower = ci[1],
ci_upper = ci[2]
)
}
# Run bootstrap
boot_results <- bootstrap_kv(data)
cat("Bootstrap results for gamma1:\n")
cat("Estimate:", boot_results$mean, "\n")
cat("SE:", boot_results$se, "\n")
cat("95% CI:", boot_results$ci_lower, "to", boot_results$ci_upper, "\n")Extensions and Future Work
Simultaneous Equations
While the current implementation focuses on triangular systems, the Klein & Vella approach could potentially be extended to simultaneous equations under additional assumptions.
Conclusion
The Klein & Vella (2010) method provides a powerful alternative to IV-based approaches for handling endogeneity through heteroskedasticity. Its control function framework is particularly valuable when:
- Traditional instruments are unavailable
- The constant correlation assumption is reasonable
- Heteroskedasticity patterns are strong
The hetid package provides both parametric and
semiparametric implementations, allowing researchers to choose the
approach that best fits their application.
See Also
- Theory and Methods - Mathematical foundations
- Getting Started - Basic Lewbel (2012) implementation
- Rigobon Method - Regime-based identification
- Prono Method - GARCH-based identification
- Package Comparison - Software validation